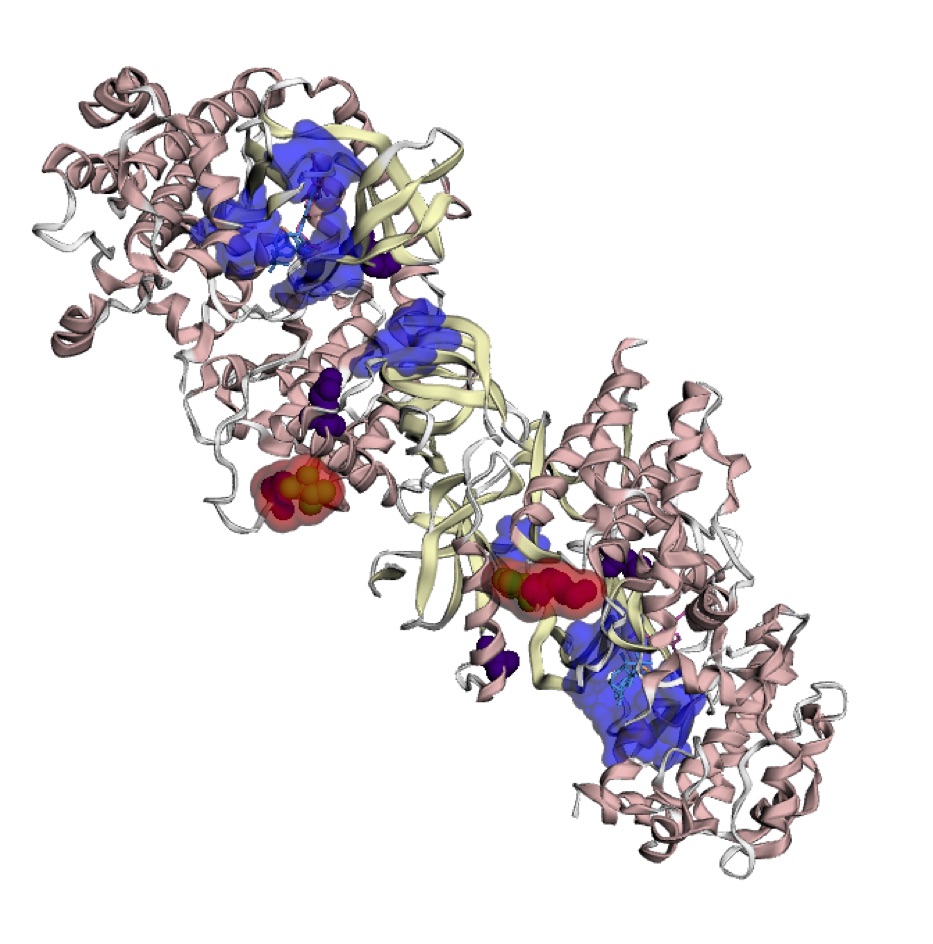
It has been hypothesized that hotspots may be cancer drug targets, biomarkers of cancer risk, and response to immunotherapy. By mapping mutations onto 3D protein structures, it is possible to identify mutational “hotspots” that are not detectable on linear amino acid sequences. As part of the TCGA Ovarian Cancer working group, we identified 3D hotspots of predicted driver missense mutations found at equivalent positions in three kinase protein families (STKs, MAPKs, and CDKs). We subsequently developed an automated method HotMAPs to analyze protein structures across the annotated exome and find statistically significant 3D hotspot regions. We mapped all missense mutations identified in 31 TCGA projects onto Protein Data Bank X-ray crystal structures plus theoretical models and applied HotMAPs. Out of >8500 tumor samples, we found that >5000 contained at least one significant missense mutation 3D hotspot, supporting the hypothesis that these events occur regularly. We also found that the percentage of samples with hotspots varied across different cancer types. Hotspot regions were shown to occur in both oncogenes (OG) and tumor suppressor genes (TSG), and we developed features and a machine learning classifier that could distinguish between OG and TSG hotspots (auROC=0.84). These results were made available through the Mutational Position Interactive Toolkit (MuPIT), our web viewer of mutation location on protein structure. Users can visualize hotspot regions found in TCGA pan-cancer analysis or for a specific cancer type.