 We have developed algorithms to reconstruct tumor clonal architectures and evolutionary trees from bulk DNA sequencing, using small somatic alterations as clonal markers. The algorithms were designed to model either single or multiple tumor samples from the same individual, either spatial or temporal. The reconstructions include estimates of the subclone proportions in each sample, which allow us to identify subclones that are expanding or contracting over space and/or time. The most expanded subclones are expected to be those with the highest fitness.
We have developed algorithms to reconstruct tumor clonal architectures and evolutionary trees from bulk DNA sequencing, using small somatic alterations as clonal markers. The algorithms were designed to model either single or multiple tumor samples from the same individual, either spatial or temporal. The reconstructions include estimates of the subclone proportions in each sample, which allow us to identify subclones that are expanding or contracting over space and/or time. The most expanded subclones are expected to be those with the highest fitness.
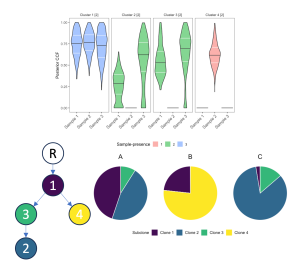
Clonal reconstruction from bulk sequence requires estimation of the prevalence of each somatic alteration in a tumor, which is not directly observable after DNA molecules are decoupled from their cell-of-origin and mixed prior to tumor sequencing. However, sequence reads can be used to cluster alterations with similar prevalence into a subclone and to infer ancestral relationships among subclones, defining the topology of an evolutionary tree. Our first method SCHISM introduced a statistical hypothesis test to quantify whether differences in prevalence were significant and a genetic algorithm to sample and score the very large space of possible trees. We have also developed a method called PICTograph that uses Bayesian hierarchical modeling for prevalence estimation and the Gabow-Myers algorithm for tree reconstruction.
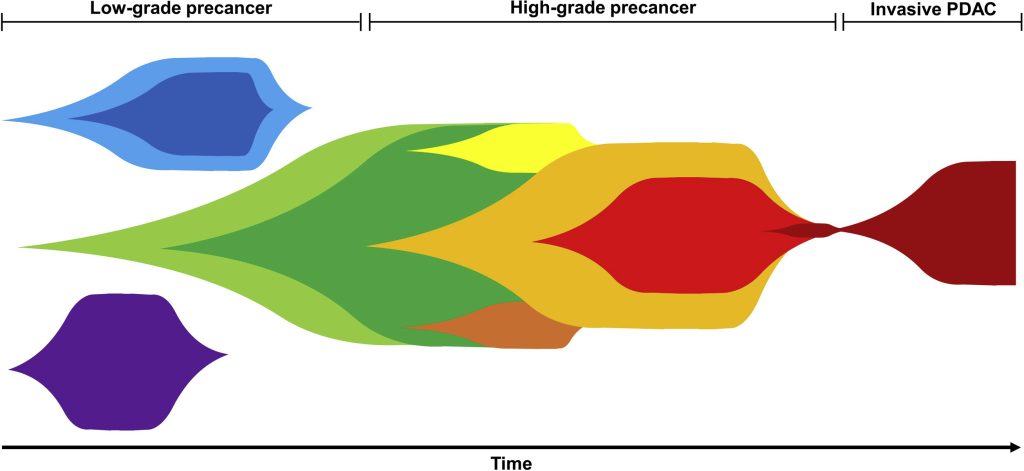
We have applied these tools in studies of high grade serous ovarian cancers (HGSOC), pancreatic cancer (PDAC) precursor lesions, pancreatic cancer in engineered mouse models, and non-small cell lung cancer tumors before and after immune checkpoint blockade (ICB) treatment (unpublished). In the HGSOC study, our modeling provided the first genetic evidence that HGSOC originates in the fallopian tube. In PDAC precursors, we found evidence that some of these neoplasms are not initiated by a single driver event but by multiple driver events that initiate competing clones, which was further supported by a single-cell DNA study. We are now developing algorithms that incorporate subclonal somatic copy number and structural alterations.


